Proteomics Facility
As part of the National Phenome Centre (NPC) at Imperial College London, we are incorporating proteomics to complement our existing lipidomics, metabolomics and genomics technology. As we develop multi-omic methods, it is a great time to discuss any proteomics that you may want to perform now or in the future. To find out more about what we are working on or to get in touch, read below!
Proteomics
Proteomics is the study of proteins. Proteins are one of the major molecular classes in the body, and are critical for controlling chemical reactions, allowing us to move, sense, taste, breath...everything! There are approximately 22,000 genes in the human genome that each contain precise instructions to make different proteins. But there are well over 1,000,000 different proteoforms in the body as they can be extensively modified to change their location, function and lifespan (turnover). Proteomics is the study of all these, to find out their biological purpose and how they perform their precise functions in health and disease. Within this facility, we focus on clinical proteomics and are developing methods for high-throughput serum proteomics, multi-omic analysis (metabolomics and genomics), targeted quantitative proteomics as well as whole proteome profiling.
Mass Spectrometry
Mass spectrometry (MS) is the principle tool of proteomics, it is an incredibly powerful technique for identifying and quantitating millions of different proteins/proteoforms. Mass spectrometry works through determining the mass of a molecule, with each molecule having a specific mass or series of masses. It can be coupled with chromatography (LC-MS) to give very accurate quantification. There are many mass spectrometry methods adjusting speed, resolution, and accuracy to the experimental requirements. MS can be set up to detect either a single protein/peptide, or thousands of different proteins in a single run.
Equipment
- M-Class UPLC (Waters) - ZenoTOF 7600 (Sciex)
- M-Class UPLC (Waters) - Synapt G2S (Waters)
- Acquity HPLC (Waters)
- Xevo G2S (Waters)
- Xevo TQ (Waters)
- Andrew+ Liquid Handling Robot (Waters)
Proteomic Profiling
ZenoTOF 7600 or Synapt G2S
Whole proteome profiling of cell lysates, tissue or bio-fluids, with or without sample pre-fractionation can be performed on our nanoLC (Acquity, Waters) coupled to either our Q-TOF (ZenoTOF 7600, Sciex) or IM-QToF instrument (Synapt G2S, Waters). The latter system features ion mobility. Acquisition can be through data-dependant (DDA) or data independent (DIA) acquisition approaches. DIA is performed through either ZenoSWATH or MSE.
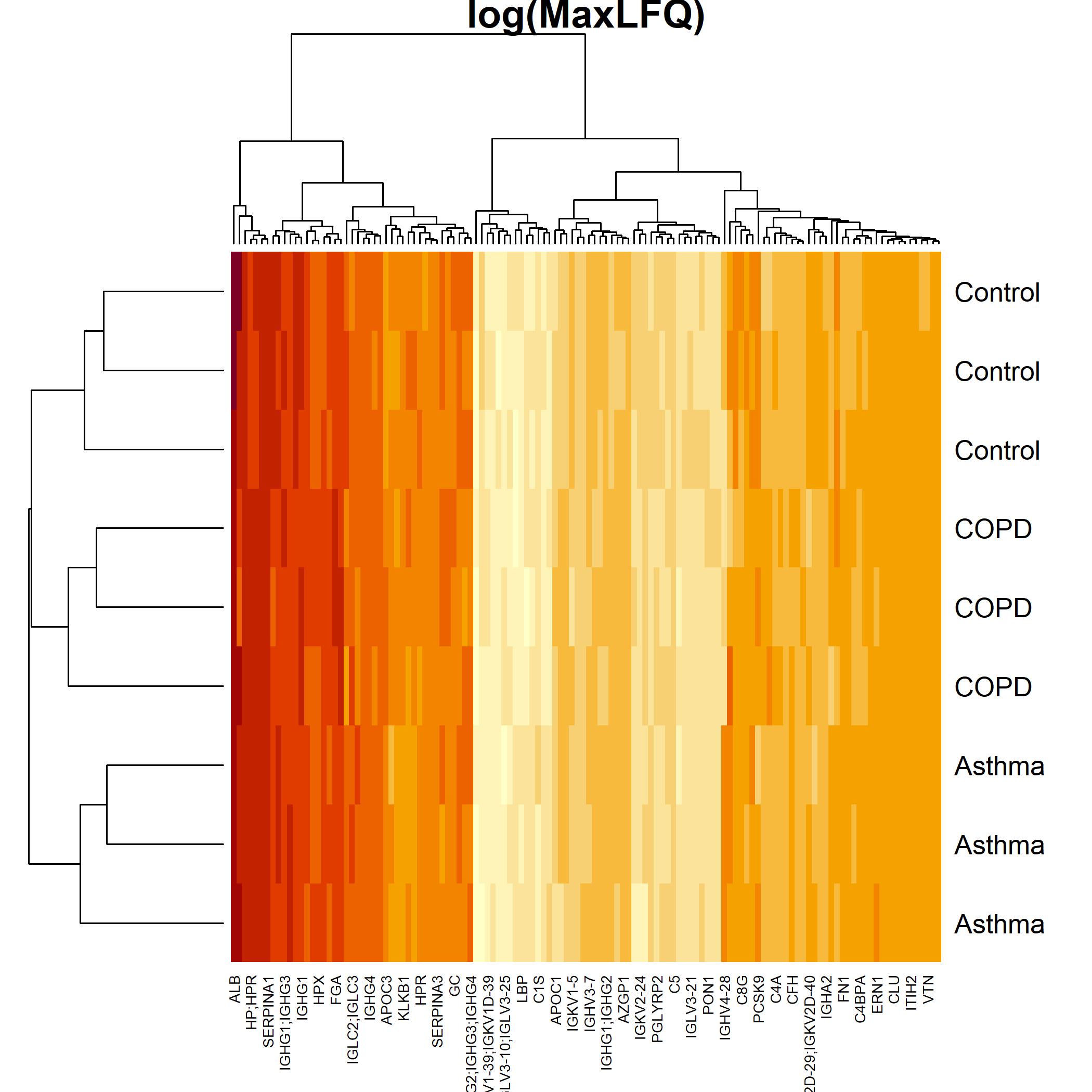
High Through-put Blood Proteomics
ZenoTOF 7600
We are setting up methods for high-throughput blood proteomics. By analysing samples in a throughput of 14 mins per sample, we are able to aquire samples on the same time frame as small-molecule profiling. This enables multi-omics profiling of the same samples without substantial lag from proteomics. Data is processed in DIA-NN with protein quantitation performed after peptide-level run-order and batch correction using the maxLFQ algorithm. We are further developing peptide-filtering approach to identify high-fidelty features for protein quantitation.
[Data in heatmap from Hughes, King and Gethings (2019) and analysed in house with DIA-NN (Demichev 2019)]
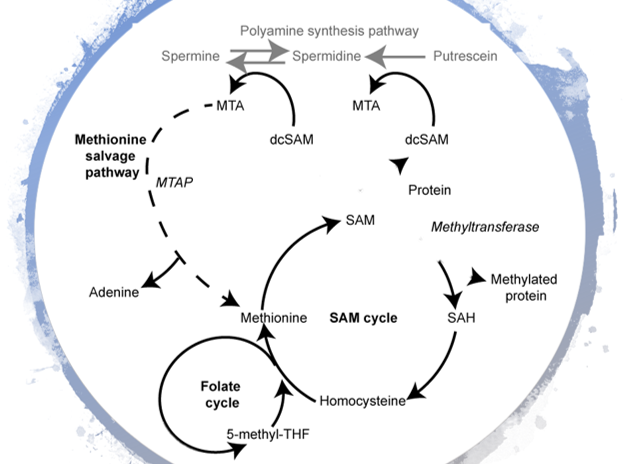
Metabo-Proteomics
The NPC has established workflows for metabolomics. To combine this with proteomics, we are developing methods for the simultaneous preparation of metabolites and proteins for quantitative analysis. This will give in depth information regarding changes to molecular pathways, mediated by proteins and metabolites within the cell. Of particular interest is the investigation of S-Adenosine-L-Methionine Cycle and Methionine Salvage Pathways.

Proteogenomics
Proteogenomics combines transcriptomic and proteomic technologies to identify splicevarients, SNPs and novel-transcriptional start sights (e.g. trascribed lnRNA, novel peptides/proteins) that would be difficult with either technology alone. Working with the genomcis facility, we are implementing experimental and analytical pipelines for to exploit protein and transcript information to discovery workflows. As we expand this technology, we will develop in house spectral libraries to increase the sensitivity of detection of different splice-forms and SNPs for future experiments.

Targeted Proteomics
MRM Assay Development
The NPC has extensive expertise in targetted assay development. We are working with a number of research groups at Imperial College London to develop targeted assays for small signalling peptides in the blood. If you are interested in developing something for your own experimental requirements get in touch!
